Functional Genomics of Maize Gametophytes
Project Description
After Meiosis, Plant Spores Develop into Multicellular Gametophytes Dependent on Their own Haploid Genomes as well as the Parent Diploid Plant
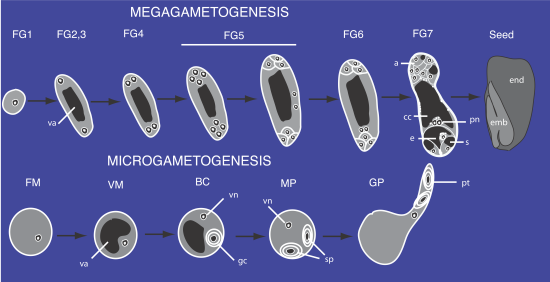
cc central cell, a antipodals, e egg, pn polar nuclei, s synergid, emb embryo, gc generative cell, end endosperm, vn vegetative nucleus, va vacuole, sp sperm cells, pt pollen tube. FG1-FG7 female gametophyte stage 1 to 7. FM free microspore, VM vacuolated microspore, BC bicellular pollen, MP mature pollen, GP germinating pollen.
Gametophyte Transcriptome Analysis
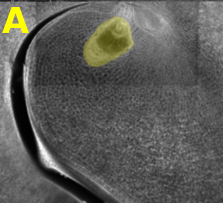
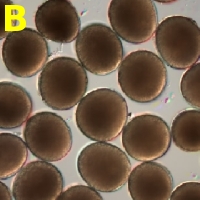
The transcriptome of mature embryo sacs and mature pollen grains are to be compared to each other and to diploid sporophyte tissue using microarrays and/or large-scale sequencing technologies. (A) Mature embryo sac in ovule. The haploid embryo sac (colored yellow) comprises only a small number of the cells, but these are much larger than and distinct from the surrounding nuclear cells. Embryo sacs are purified in two ways: 1) using 15micrometre paraffin sections, each embryo sac will be contained in 4 or 5 sections, allowing us to collect pure haploid tissue by LCM; (2) embryo sacs are to be isolated using dissecting needles from ovules treated with cell wall degrading enzymes. (B) Mature pollen grains.
The female and male gametophytes make up the haploid phase of the angiosperm life cycle, which immediately precedes formation of the seed (embryo and endosperm). Although gametophytes are small and undergo few cell divisions, they are crucial for producing the next generation.
However, relatively little is known about the genetic and cellular mechanisms underlying gametophyte function and development, primarily because mutations that are deleterious to these haploid tissues are difficult to recover and maintain. This project seeks to overcome this limitation using genetic tools unique to the model crop Zea mays (maize) to accomplish a genomic-scale investigation of gametophytically-required genes. Trisomic stocks that transmit duplicate chromosomal regions through the gametophyte will be used to screen for Ac transposon-tagged gametophyte-lethal mutations. These mutations will be phenotypically characterized in both gametophytes, and their corresponding DNA sequences identified. Verification of the identity of a select group of candidate genes will be accomplished using multiple alleles and RNA expression analyses. Complementary aims in expression profiling of both gametophytes, and in bioinformatics to identify orthologous genes in other plant models and to integrate the multiple types of data generated by the project, will allow an assessment of the genetic basis of gametophyte function. Finally, the ability to predict gametophytic functions across species boundaries will be tested using RNAi to target select genes in the dicot model Arabidopsis thaliana. An understanding of the genetic basis of gametophyte function will have important implications for agriculture, as gametophytes are central to plant reproduction.
Broader Impacts
This project is relevant to many agricultural objectives seeking to influence plant reproduction - for example, controlling pollen fertility for hybrid seed production, limiting pollen-mediated transgene flow, and inducing apomixis. Because the tools and stocks created, and sequences generated, will be freely available to the scientific community, the project will enable other researchers to better investigate gametophytes through creation of a gametophyte-specific sequence-indexed mutant collection. The outreach component of the project will help train 10 undergraduates to serve as science mentors for K-12 students, through exposing them to plant science research. In partnership with Stanford University's Haas Center for Public Service, these undergraduate mentors will be placed in communities with large populations from under-represented groups, and help develop science experiences for K-12 students. In addition, the project will train two postdoctoral researchers and a graduate student in genomic-scale approaches to plant biology, and will introduce undergraduates and high school students to genomic science.
Access to Project Outcomes
All stocks created by the project will be deposited in the Maize Coop Stock Center. All Ac transposon flanking sequences will be searchable by BLAST, both at Plant Genome Database and at this project's web site, maizegametophyte.org. Phenotypic and expression data will also be accessible at ZGamDb. For long-term storage and broad community access, phenotypic data will be incorporated into MaizeGDB, and expression data will be deposited at the Gene Expression Omnibus and Plant Expression Database.
Project Support:
This project is funded by a 2007 award from the National Science Foundation, Plant Genome Research Program, Award #0701731








